Develop biological databases and bioinformatics tools to support biomedical research and analytical techniques.
HuMemprotDB
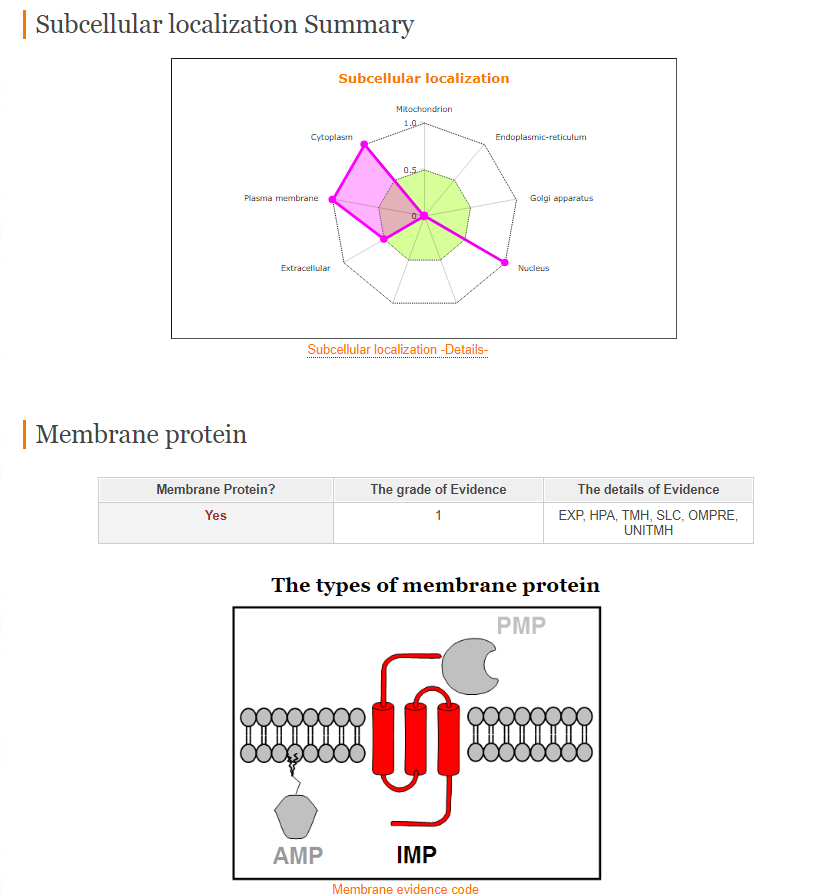
HuMemProtDB is a curated, searchable, relational database on integral, peripheral and anchored membrane proteins for human. Protein sequences are collected from UniPort, International Protein Index (IPI) and Human Protein Reference Database (HPRD).
HuMemProtDB provides the following analysis:
1) Membrane protein analysis
2) Structural analysis
3) Subcellular localization analysis
4) Biomarker analysis (Functional analysis)

iHPDM
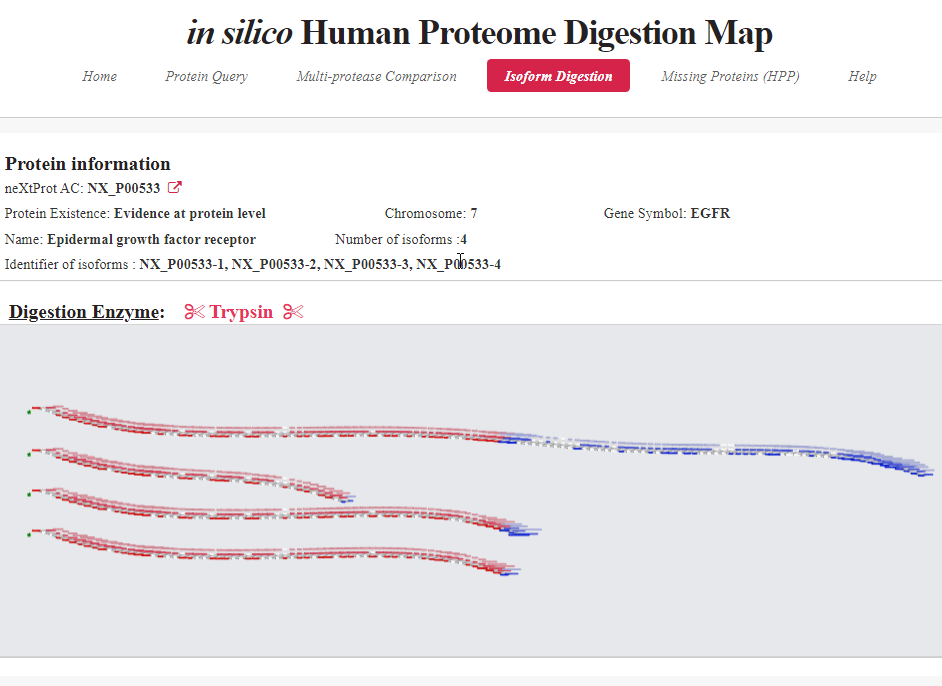
iHPDM contains a comprehensive peptide database of the human proteins in neXtProt digested by 15 protease combinations of single and two proteases. Also, it provides convenient functions and graphical visualizations for users to examine and compare the digestion results of different proteases.
iHPDM provide the three main functions for protease selection in shotgun proteomics experiments to identify missing proteins, protein isoforms, and single amino acid variant peptides.
1) Protein Query
2) Multiprotease Comparison
3) Isoform Digestion

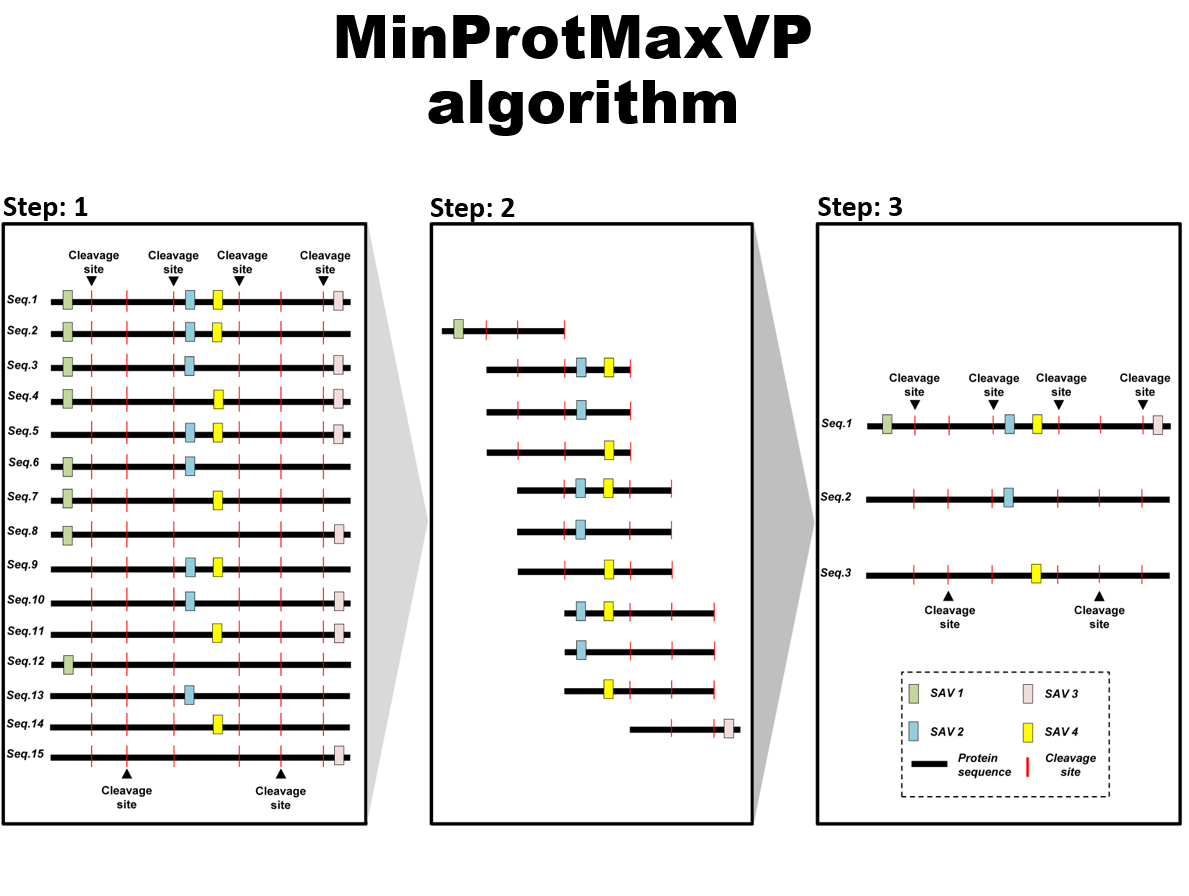
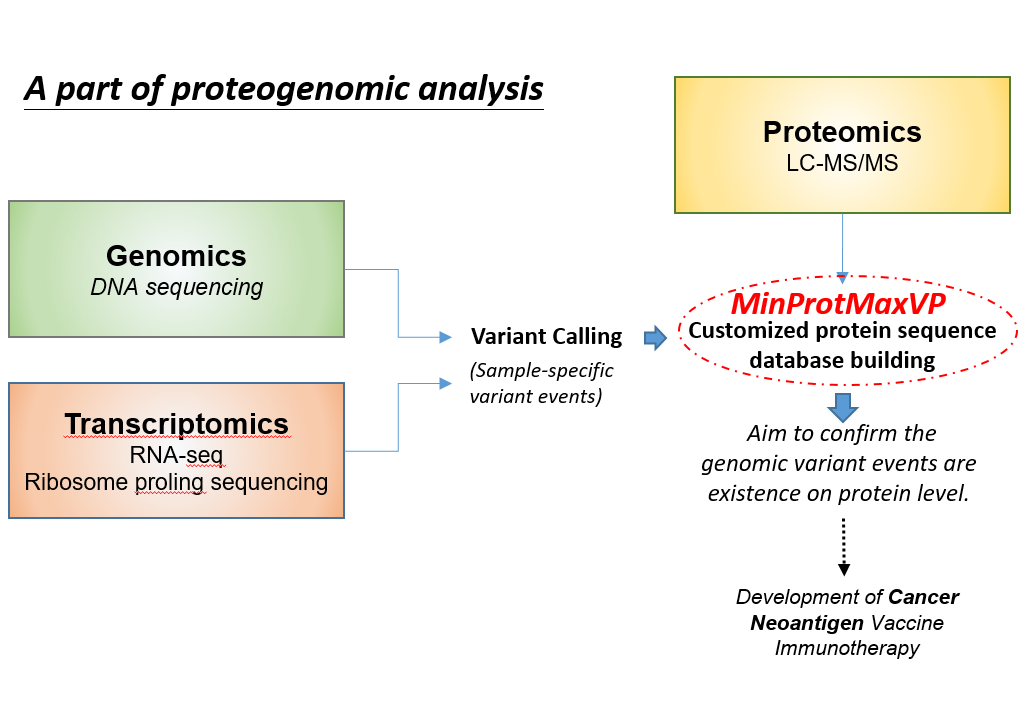
MinProtMaxVP
MinProtMaxVP, a novel approach to generate SAV-harboring protein sequences for constructing a customized protein sequence database, which is used in database searching for variant peptide identification.
gDecoy
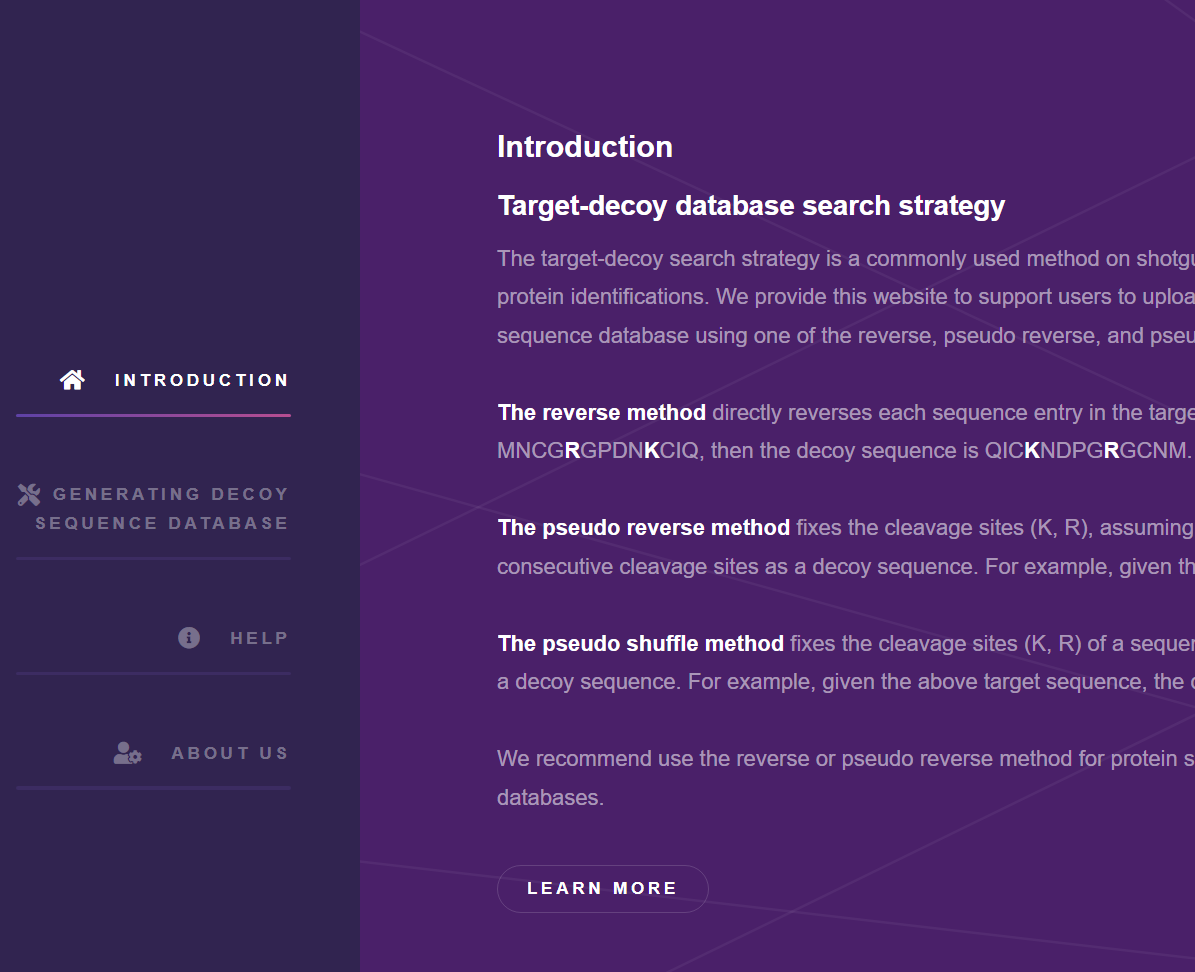
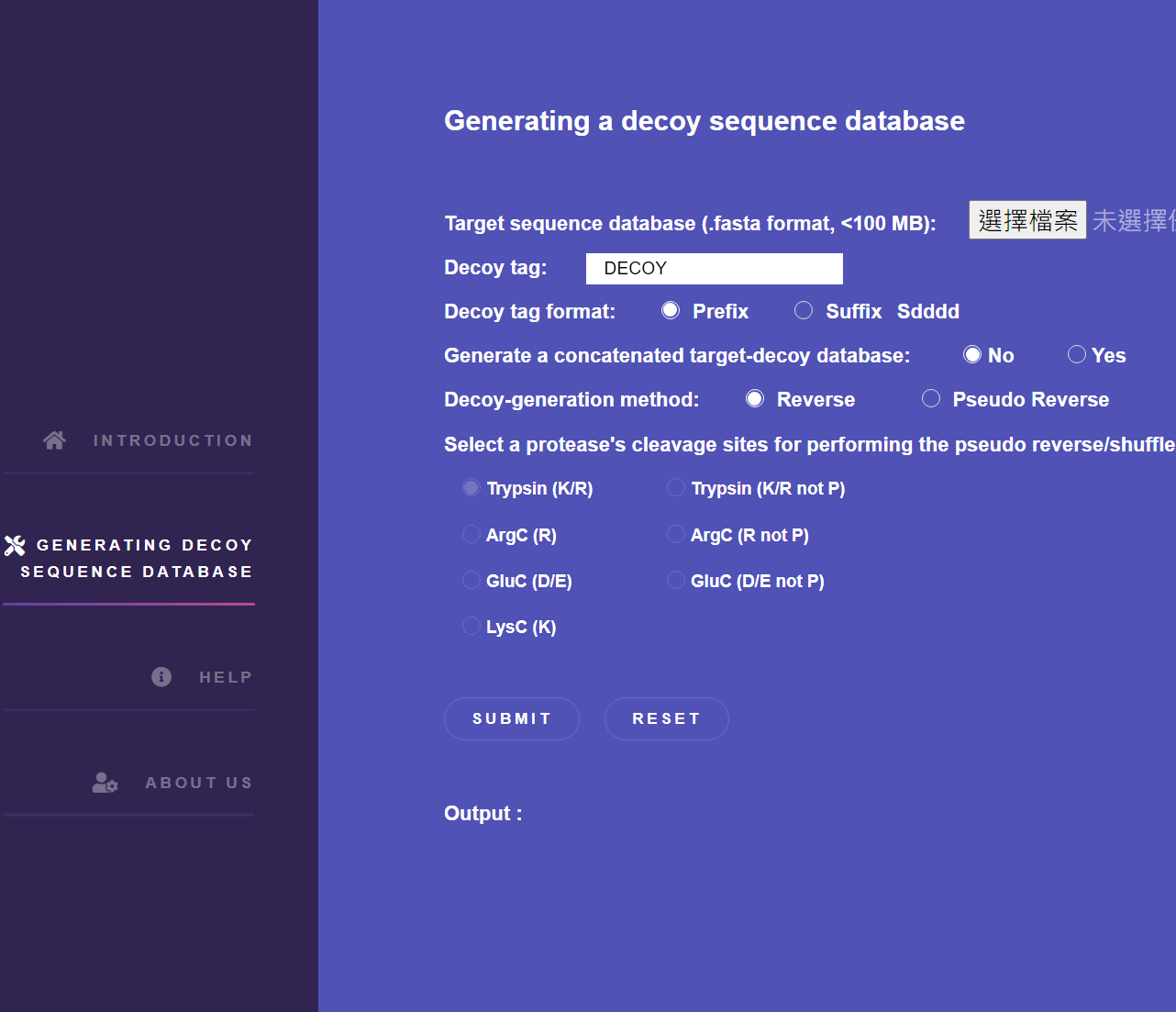
A web-based tool is provided to construct a suitable decoy database for both the protein-based and the peptide-based sequence databases used in target-decoy searches. It allows users to select a decoy generation method and the enzyme used to generate decoy peptides from in silico digested peptides when using the pseudo reverse or the pseudo shuffle methods.
 (Taiwan)
(Taiwan)